V(D)J recombinations in lymphocytes are essential for immunological diversity. They are also useful markers of pathologies, and in leukemia, are used to quantify the minimal residual disease during patient follow-up. High-throughput sequencing (NGS/HTS) now enables the deep sequencing of lymphoid populations. With dedicated Rep-Seq bioinformatics methods and software, clinicians and researchers can now qualify and sometimes quantify the heterogeneity of these lymphoid populations.
This workshop was intended for users and developers of Vidjil, to share experience, protocols and results, to raise questions and decide development priorities, and possibly to set up new collaborations between biologists and bioinformaticians. The workshop included two invited talks, hematology, immunology and bioinformatics talks from the users and developers of Vidjil, and practical hands-on sessions on participant data. The workshop gathered 34 scientists from 5 countries.
Program
The workshop will start on the Tuesday at 9:30 and end on the Wednesday at 16:00. For the practical sessions, the participants are invited to bring a laptop and some of their NGS data (fasta / fastq / fastq.gz files). The data can also be uploaded into user accounts on app.vidjil.org before Sunday 13 March. There will be no internet connection in the workshop rooms, but all the data uploaded before March 13 will be available on a local server. We will also provide test data for the people who do not currently have their own NGS Rep-Seq data.
Tuesday 15 March
| 9:30 – 9:50 | Registration, coffee |
| 9:50 – 10:00 | Introduction |
| 10:00 – 11:00 | Invited talk: John Moppett
Evolving the role of MRD in the management of childhood leukaemia University Hospitals Bristol, NHS (UK) |
| 11:00 – 12:30 | Diagnosis and MRD for Leukemia and Lymphoma
|
| 12:30 – 14:00 | Lunch |
| 14:00 – 15:30 | Developer talks: Bioinformatics and algorithmics
|
| 15:30 – 16:00 | Coffee Break |
| 16:00 – 17:30 | Practical session 1 |
| 20:30 | Dinner, Art-Déco restaurant La Paix in Lille |
Wednesday 16 March
| 9:00 – 10:00 | Invited talk: Mikhail Shugay
A general-purpose framework for immune repertoire sequencing: applications to aging and autoimmune disorder studies Genomics of Adaptive Immunity Lab Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Moscow (Russia) |
| 10:00 – 10:30 | Coffee Break |
| 10:30 – 12:00 | Clonality and Repertoire Analysis in Hematology and Immunology
|
| 12:00 – 13:30 | Lunch |
| 13:30 – 15:00 | Practical session 2 |
| 15:00 – 15:20 | Coffee Break |
| 15:20 – 16:00 | Round Table Discussion, with Martin Figeac, Mathieu Giraud, John Moppett, Mikaël Salson and Mikhail Shugay. Discussion on library preparation, sequencing instruments and protocols, bioinformatics analysis. Perspectives and possible collaborations. |
Practical information
Lille is a lovely city at the heart of French Flanders. With TGV, Lille is 1h from Paris, 1h20 min from London and 35 min from Brussels.
List of hotels with available rooms
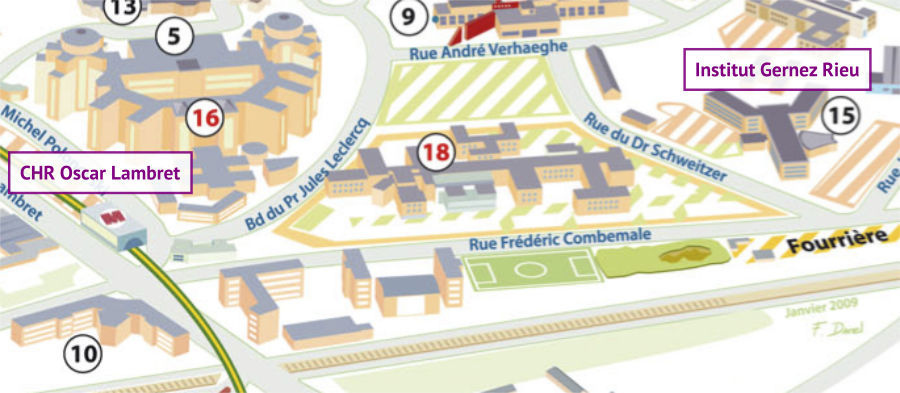
The workshop will take place at Institut Gernez Rieu (IGR), in the heart of the Lille Hospital, at 15 minutes from Lille center. From Lille center, take the metro line 1 (yellow), direction CHR B Calmette. Stop at the CHR Oscar Lambret station. The IGR building is 600 meters from the metro station.
Organizers
The workshop is organized by Mathieu Giraud, Ryan Herbert, Tatiana Rocher, Mikaël Salson, Maximilien Vanleene and Aurélie Wieczorek, from CRIStAL, Inria Lille and SIRIC OncoLille.




